Graph Neural Networks for Unsupervised Domain Adaptation of Histopathological ImageAnalytics
Abstract
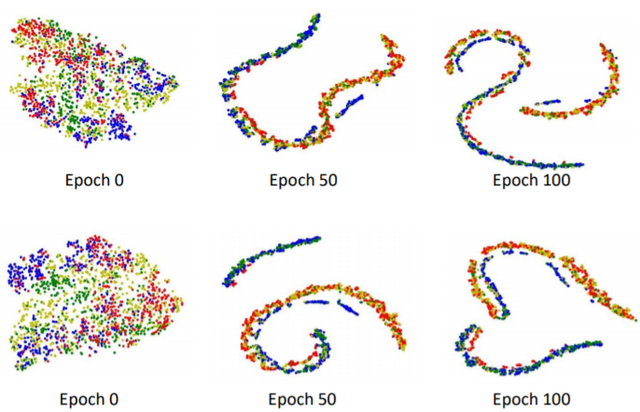
Annotating histopathological images is a time-consuming andlabor-intensive process, which requires broad-certificated pathologistscarefully examining large-scale whole-slide images from cells to tissues.Recent frontiers of transfer learning techniques have been widely investi-gated for image understanding tasks with limited annotations. However,when applied for the analytics of histology images, few of them can effec-tively avoid the performance degradation caused by the domain discrep-ancy between the source training dataset and the target dataset, suchas different tissues, staining appearances, and imaging devices. To thisend, we present a novel method for the unsupervised domain adaptationin histopathological image analysis, based on a backbone for embeddinginput images into a feature space, and a graph neural layer for propa-gating the supervision signals of images with labels. The graph model isset up by connecting every image with its close neighbors in the embed-ded feature space. Then graph neural network is employed to synthesizenew feature representation from every image. During the training stage,target samples with confident inferences are dynamically allocated withpseudo labels. The cross-entropy loss function is used to constrain thepredictions of source samples with manually marked labels and targetsamples with pseudo labels. Furthermore, the maximum mean diversityis adopted to facilitate the extraction of domain-invariant feature repre-sentations, and contrastive learning is exploited to enhance the categorydiscrimination of learned features. In experiments of the unsupervised do-main adaptation for histopathological image classification, our methodachieves state-of-the-art performance on four public datasets.