Deep Feature Representation for The Computational Analytics of 3D Neuronal Morphology
Abstract
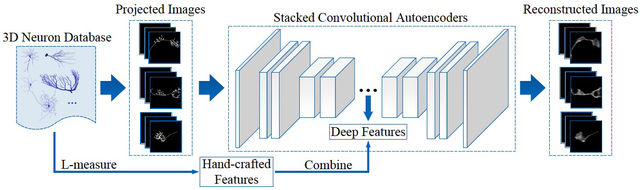
With the ever-increasing number of digitally reconstructed neurons, computational analytics of 3D neuronal morphology has become a new avenue to understand neuroanatomical structures and functional properties. However, traditional methods cannot well identify and represent neuronal morphology, especially when tackling large-scale and diverse neuron datasets. In this paper, we propose a deep learning based framework for the representation of 3D neuron morphology. At first, considering the neuronal tree-structures are usually very sparse in 3D space, we project each 3D neuron into 2D images with three angles of view. The projective strategy can preserve the spatial morphologies from the original data. Subsequently, as there are no sufficient annotations for each neuron, we introduce an unsupervised deep neural network to automatically learn neuron features from the projected 2D images. The deep features are then combined with traditional features for accurate neuron representation. Experimental results validate the effectiveness of our method by searching similar samples on a public database including 58; 000 neurons. Moreover, we demonstrate that the traditional hand-crafted features are complementary with deep features in the representation of 3D neuron morphology.